kwcoco.metrics.confusion_vectors module¶
Classes that store raw confusion vectors, which can be accumulated into confusion measures.
- class kwcoco.metrics.confusion_vectors.ConfusionVectors(data, classes, probs=None)[source]¶
Bases:
NiceReprStores information used to construct a confusion matrix. This includes corresponding vectors of predicted labels, true labels, sample weights, etc…
- Variables:
data (kwarray.DataFrameArray) – should at least have keys true, pred, weight
classes (Sequence | CategoryTree) – list of category names or category graph
probs (ndarray | None) – probabilities for each class
Example
>>> # xdoctest: IGNORE_WANT >>> # xdoctest: +REQUIRES(module:pandas) >>> from kwcoco.metrics import DetectionMetrics >>> dmet = DetectionMetrics.demo( >>> nimgs=10, nboxes=(0, 10), n_fp=(0, 1), classes=3) >>> cfsn_vecs = dmet.confusion_vectors() >>> print(cfsn_vecs.data._pandas()) pred true score weight iou txs pxs gid 0 2 2 10.0000 1.0000 1.0000 0 4 0 1 2 2 7.5025 1.0000 1.0000 1 3 0 2 1 1 5.0050 1.0000 1.0000 2 2 0 3 3 -1 2.5075 1.0000 -1.0000 -1 1 0 4 2 -1 0.0100 1.0000 -1.0000 -1 0 0 5 -1 2 0.0000 1.0000 -1.0000 3 -1 0 6 -1 2 0.0000 1.0000 -1.0000 4 -1 0 7 2 2 10.0000 1.0000 1.0000 0 5 1 8 2 2 8.0020 1.0000 1.0000 1 4 1 9 1 1 6.0040 1.0000 1.0000 2 3 1 .. ... ... ... ... ... ... ... ... 62 -1 2 0.0000 1.0000 -1.0000 7 -1 7 63 -1 3 0.0000 1.0000 -1.0000 8 -1 7 64 -1 1 0.0000 1.0000 -1.0000 9 -1 7 65 1 -1 10.0000 1.0000 -1.0000 -1 0 8 66 1 1 0.0100 1.0000 1.0000 0 1 8 67 3 -1 10.0000 1.0000 -1.0000 -1 3 9 68 2 2 6.6700 1.0000 1.0000 0 2 9 69 2 2 3.3400 1.0000 1.0000 1 1 9 70 3 -1 0.0100 1.0000 -1.0000 -1 0 9 71 -1 2 0.0000 1.0000 -1.0000 2 -1 9
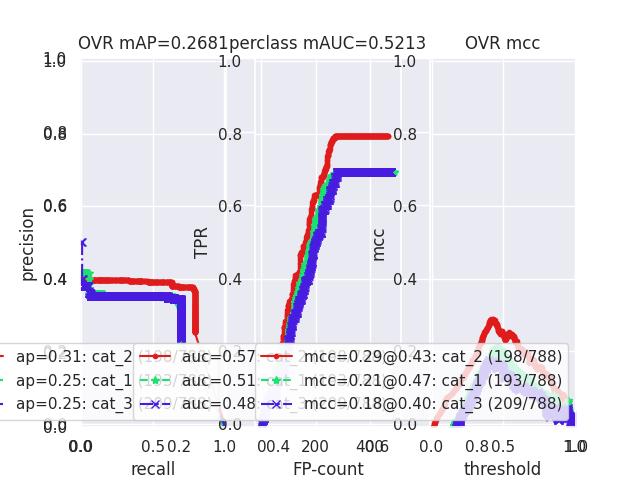
>>> # xdoctest: +REQUIRES(--show) >>> # xdoctest: +REQUIRES(module:pandas) >>> import kwplot >>> kwplot.autompl() >>> from kwcoco.metrics.confusion_vectors import ConfusionVectors >>> cfsn_vecs = ConfusionVectors.demo( >>> nimgs=128, nboxes=(0, 10), n_fp=(0, 3), n_fn=(0, 3), classes=3) >>> cx_to_binvecs = cfsn_vecs.binarize_ovr() >>> measures = cx_to_binvecs.measures()['perclass'] >>> print('measures = {!r}'.format(measures)) measures = <PerClass_Measures({ 'cat_1': <Measures({'ap': 0.227, 'auc': 0.507, 'catname': cat_1, 'max_f1': f1=0.45@0.47, 'nsupport': 788.000})>, 'cat_2': <Measures({'ap': 0.288, 'auc': 0.572, 'catname': cat_2, 'max_f1': f1=0.51@0.43, 'nsupport': 788.000})>, 'cat_3': <Measures({'ap': 0.225, 'auc': 0.484, 'catname': cat_3, 'max_f1': f1=0.46@0.40, 'nsupport': 788.000})>, }) at 0x7facf77bdfd0> >>> kwplot.figure(fnum=1, doclf=True) >>> measures.draw(key='pr', fnum=1, pnum=(1, 3, 1)) >>> measures.draw(key='roc', fnum=1, pnum=(1, 3, 2)) >>> measures.draw(key='mcc', fnum=1, pnum=(1, 3, 3)) ...
- classmethod demo(**kw)[source]¶
- Parameters:
**kwargs – See
kwcoco.metrics.DetectionMetrics.demo()- Returns:
ConfusionVectors
Example
>>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> cfsn_vecs = ConfusionVectors.demo() >>> print('cfsn_vecs = {!r}'.format(cfsn_vecs)) >>> cx_to_binvecs = cfsn_vecs.binarize_ovr() >>> print('cx_to_binvecs = {!r}'.format(cx_to_binvecs))
- classmethod from_arrays(true, pred=None, score=None, weight=None, probs=None, classes=None)[source]¶
Construct confusion vector data structure from component arrays
Example
>>> # xdoctest: +REQUIRES(module:pandas) >>> import kwarray >>> classes = ['person', 'vehicle', 'object'] >>> rng = kwarray.ensure_rng(0) >>> true = (rng.rand(10) * len(classes)).astype(int) >>> probs = rng.rand(len(true), len(classes)) >>> cfsn_vecs = ConfusionVectors.from_arrays(true=true, probs=probs, classes=classes) >>> cfsn_vecs.confusion_matrix() pred person vehicle object real person 0 0 0 vehicle 2 4 1 object 2 1 0
- confusion_matrix(compress=False)[source]¶
Builds a confusion matrix from the confusion vectors.
- Parameters:
compress (bool, default=False) – if True removes rows / columns with no entries
- Returns:
- cmthe labeled confusion matrix
- (Note: we should write a efficient replacement for
this use case. #remove_pandas)
- Return type:
pd.DataFrame
CommandLine
xdoctest -m kwcoco.metrics.confusion_vectors ConfusionVectors.confusion_matrix
Example
>>> # xdoctest: +REQUIRES(module:pandas) >>> from kwcoco.metrics import DetectionMetrics >>> dmet = DetectionMetrics.demo( >>> nimgs=10, nboxes=(0, 10), n_fp=(0, 1), n_fn=(0, 1), >>> classes=3, cls_noise=.2, newstyle=False) >>> cfsn_vecs = dmet.confusion_vectors() >>> cm = cfsn_vecs.confusion_matrix() ... >>> print(cm.to_string(float_format=lambda x: '%.2f' % x)) pred background cat_1 cat_2 cat_3 real background 0.00 1.00 2.00 3.00 cat_1 3.00 12.00 0.00 0.00 cat_2 3.00 0.00 14.00 0.00 cat_3 2.00 0.00 0.00 17.00
- binarize_classless(negative_classes=None)[source]¶
Creates a binary representation useful for measuring the performance of detectors. It is assumed that scores of “positive” classes should be high and “negative” clases should be low.
- Parameters:
negative_classes (List[str | int] | None) – list of negative class names or idxs, by default chooses any class with a true class index of -1. These classes should ideally have low scores.
- Returns:
BinaryConfusionVectors
Note
The “classlessness” of this depends on the compat=”all” argument being used when constructing confusion vectors, otherwise it becomes something like a macro-average because the class information was used in deciding which true and predicted boxes were allowed to match.
Example
>>> from kwcoco.metrics import DetectionMetrics >>> dmet = DetectionMetrics.demo( >>> nimgs=10, nboxes=(0, 10), n_fp=(0, 1), n_fn=(0, 1), classes=3) >>> cfsn_vecs = dmet.confusion_vectors() >>> class_idxs = list(dmet.classes.node_to_idx.values()) >>> binvecs = cfsn_vecs.binarize_classless()
- binarize_ovr(mode=1, keyby='name', ignore_classes={'ignore'}, approx=False)[source]¶
Transforms cfsn_vecs into one-vs-rest BinaryConfusionVectors for each category.
- Parameters:
mode (int, default=1) – 0 for heirarchy aware or 1 for voc like. MODE 0 IS PROBABLY BROKEN
keyby (int | str) – can be cx or name
ignore_classes (Set[str]) – category names to ignore
approx (bool, default=0) – if True try and approximate missing scores otherwise assume they are irrecoverable and use -inf
- Returns:
- which behaves like
Dict[int, BinaryConfusionVectors]: cx_to_binvecs
- Return type:
Example
>>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> cfsn_vecs = ConfusionVectors.demo() >>> print('cfsn_vecs = {!r}'.format(cfsn_vecs)) >>> catname_to_binvecs = cfsn_vecs.binarize_ovr(keyby='name') >>> print('catname_to_binvecs = {!r}'.format(catname_to_binvecs))
cfsn_vecs.data.pandas() catname_to_binvecs.cx_to_binvecs[‘class_1’].data.pandas()
Note
- class kwcoco.metrics.confusion_vectors.OneVsRestConfusionVectors(cx_to_binvecs, classes)[source]¶
Bases:
NiceReprContainer for multiple one-vs-rest binary confusion vectors
- Variables:
cx_to_binvecs –
classes –
Example
>>> from kwcoco.metrics import DetectionMetrics >>> dmet = DetectionMetrics.demo( >>> nimgs=10, nboxes=(0, 10), n_fp=(0, 1), classes=3) >>> cfsn_vecs = dmet.confusion_vectors() >>> self = cfsn_vecs.binarize_ovr(keyby='name') >>> print('self = {!r}'.format(self))
- classmethod demo()[source]¶
- Parameters:
**kwargs – See
kwcoco.metrics.DetectionMetrics.demo()- Returns:
ConfusionVectors
- measures(stabalize_thresh=7, fp_cutoff=None, monotonic_ppv=True, ap_method='pycocotools')[source]¶
Creates binary confusion measures for every one-versus-rest category.
- Parameters:
stabalize_thresh (int) – if fewer than this many data points inserts dummy stabilization data so curves can still be drawn. Default to 7.
fp_cutoff (int | None) – maximum number of false positives in the truncated roc curves. The default
Noneis equivalent tofloat('inf')monotonic_ppv (bool) – if True ensures that precision is always increasing as recall decreases. This is done in pycocotools scoring, but I’m not sure its a good idea. Default to True.
Example
>>> self = OneVsRestConfusionVectors.demo() >>> thresh_result = self.measures()['perclass']
- class kwcoco.metrics.confusion_vectors.BinaryConfusionVectors(data, cx=None, classes=None)[source]¶
Bases:
NiceReprStores information about a binary classification problem. This is always with respect to a specific class, which is given by cx and classes.
- The data DataFrameArray must contain
is_true - if the row is an instance of class classes[cx] pred_score - the predicted probability of class classes[cx], and weight - sample weight of the example
Example
>>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> self = BinaryConfusionVectors.demo(n=10) >>> print('self = {!r}'.format(self)) >>> print('measures = {}'.format(ub.urepr(self.measures())))
>>> self = BinaryConfusionVectors.demo(n=0) >>> print('measures = {}'.format(ub.urepr(self.measures())))
>>> self = BinaryConfusionVectors.demo(n=1) >>> print('measures = {}'.format(ub.urepr(self.measures())))
>>> self = BinaryConfusionVectors.demo(n=2) >>> print('measures = {}'.format(ub.urepr(self.measures())))
- classmethod demo(n=10, p_true=0.5, p_error=0.2, p_miss=0.0, rng=None)[source]¶
Create random data for tests
- Parameters:
n (int) – number of rows
p_true (float) – fraction of real positive cases
p_error (float) – probability of making a recoverable mistake
p_miss (float) – probability of making a unrecoverable mistake
rng (int | RandomState | None) – random seed / state
- Returns:
BinaryConfusionVectors
Example
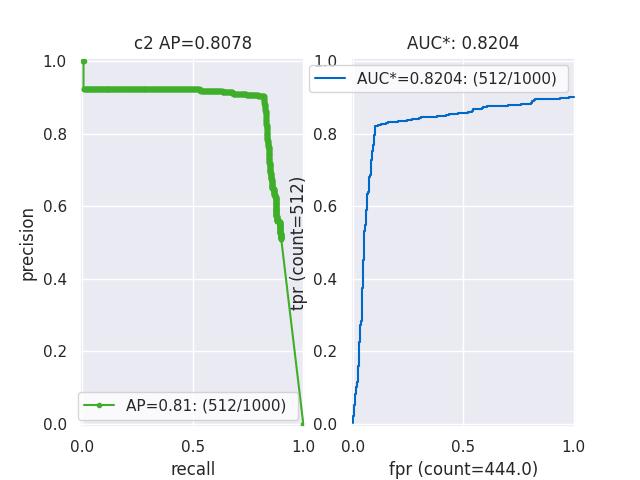
>>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> cfsn = BinaryConfusionVectors.demo(n=1000, p_error=0.1, p_miss=0.1) >>> measures = cfsn.measures() >>> print('measures = {}'.format(ub.urepr(measures, nl=1))) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.figure(fnum=1, pnum=(1, 2, 1)) >>> measures.draw('pr') >>> kwplot.figure(fnum=1, pnum=(1, 2, 2)) >>> measures.draw('roc')
- property catname¶
- measures(stabalize_thresh=7, fp_cutoff=None, monotonic_ppv=True, ap_method='pycocotools')[source]¶
Get statistics (F1, G1, MCC) versus thresholds
- Parameters:
stabalize_thresh (int, default=7) – if fewer than this many data points inserts dummy stabalization data so curves can still be drawn.
fp_cutoff (int | None) – maximum number of false positives in the truncated roc curves. The default of
Noneis equivalent tofloat('inf')monotonic_ppv (bool) – if True ensures that precision is always increasing as recall decreases. This is done in pycocotools scoring, but I’m not sure its a good idea.
Example
>>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> self = BinaryConfusionVectors.demo(n=0) >>> print('measures = {}'.format(ub.urepr(self.measures()))) >>> self = BinaryConfusionVectors.demo(n=1, p_true=0.5, p_error=0.5) >>> print('measures = {}'.format(ub.urepr(self.measures()))) >>> self = BinaryConfusionVectors.demo(n=3, p_true=0.5, p_error=0.5) >>> print('measures = {}'.format(ub.urepr(self.measures())))
>>> self = BinaryConfusionVectors.demo(n=100, p_true=0.5, p_error=0.5, p_miss=0.3) >>> print('measures = {}'.format(ub.urepr(self.measures()))) >>> print('measures = {}'.format(ub.urepr(ub.odict(self.measures()))))
References
https://en.wikipedia.org/wiki/Confusion_matrix https://en.wikipedia.org/wiki/Precision_and_recall https://en.wikipedia.org/wiki/Matthews_correlation_coefficient
- _binary_clf_curves(stabalize_thresh=7, fp_cutoff=None)[source]¶
Compute TP, FP, TN, and FN counts for this binary confusion vector.
Code common to ROC, PR, and threshold measures, computes the elements of the binary confusion matrix at all relevant operating point thresholds.
- Parameters:
stabalize_thresh (int) – if fewer than this many data points insert stabalization data.
fp_cutoff (int | None) – maximum number of false positives
Example
>>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> self = BinaryConfusionVectors.demo(n=1, p_true=0.5, p_error=0.5) >>> self._binary_clf_curves()
>>> self = BinaryConfusionVectors.demo(n=0, p_true=0.5, p_error=0.5) >>> self._binary_clf_curves()
>>> self = BinaryConfusionVectors.demo(n=100, p_true=0.5, p_error=0.5) >>> self._binary_clf_curves()
- _3dplot()[source]¶
Example
>>> # xdoctest: +REQUIRES(module:kwplot) >>> # xdoctest: +REQUIRES(module:pandas) >>> from kwcoco.metrics.confusion_vectors import * # NOQA >>> from kwcoco.metrics.detect_metrics import DetectionMetrics >>> dmet = DetectionMetrics.demo( >>> n_fp=(0, 1), n_fn=(0, 2), nimgs=256, nboxes=(0, 10), >>> bbox_noise=10, >>> classes=1) >>> cfsn_vecs = dmet.confusion_vectors() >>> self = bin_cfsn = cfsn_vecs.binarize_classless() >>> #dmet.summarize(plot=True) >>> import kwplot >>> kwplot.autompl() >>> kwplot.figure(fnum=3) >>> self._3dplot()
- kwcoco.metrics.confusion_vectors._stabalize_data(y_true, y_score, sample_weight, npad=7)[source]¶
Adds ideally calibrated dummy values to curves with few positive examples. This acts somewhat like a Bayesian prior and smooths out the curve.
Example
y_score = np.array([0.5, 0.6]) y_true = np.array([1, 1]) sample_weight = np.array([1, 1]) npad = 7 _stabalize_data(y_true, y_score, sample_weight, npad=npad)